SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes
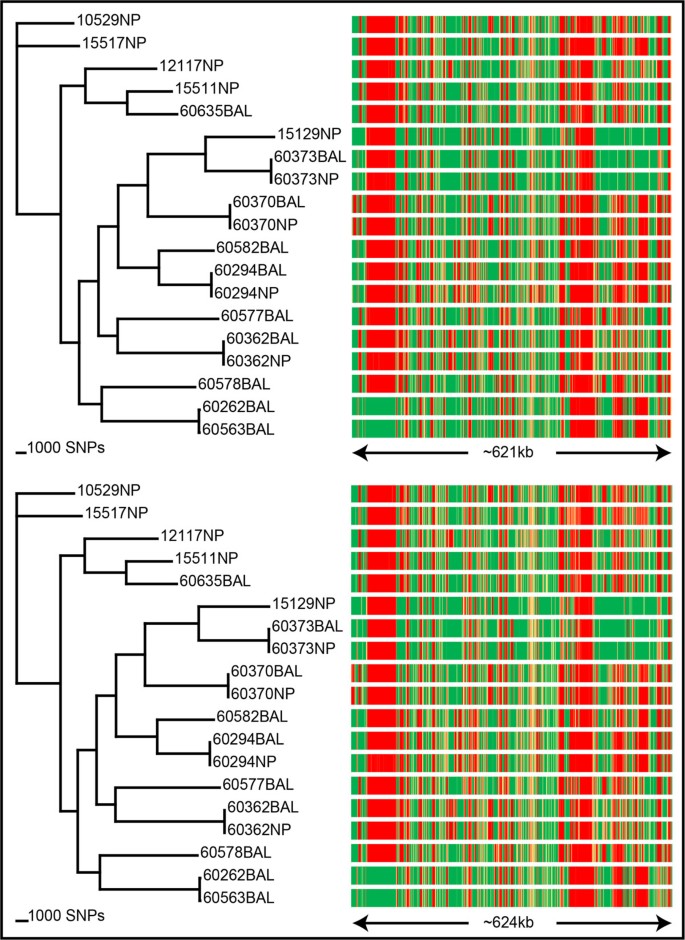

Comparative genomics and antimicrobial resistance profiling of Elizabethkingia isolates reveals nosocomial transmission and in vitro susceptibility to fluoroquinolones, tetracyclines and trimethoprim-sulfamethoxazole

VDAP-GUI: a user-friendly pipeline for variant discovery and annotation of raw next-generation sequencing data

Using whole-genome sequencing and a pentaplex real-time PCR to characterize third-generation cephalosporin-resistant Enterobacteriaceae from Southeast Queensland, Australia
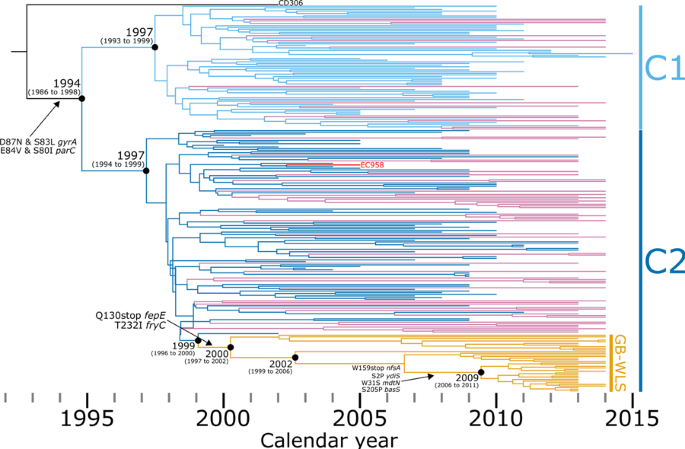
Genomic epidemiology reveals geographical clustering of multidrug-resistant Escherichia coli ST131 associated with bacteraemia in Wales

Comparative genomics of Nocardia seriolae reveals recent importation and subsequent widespread dissemination in mariculture farms in South Central Coast, Vietnam
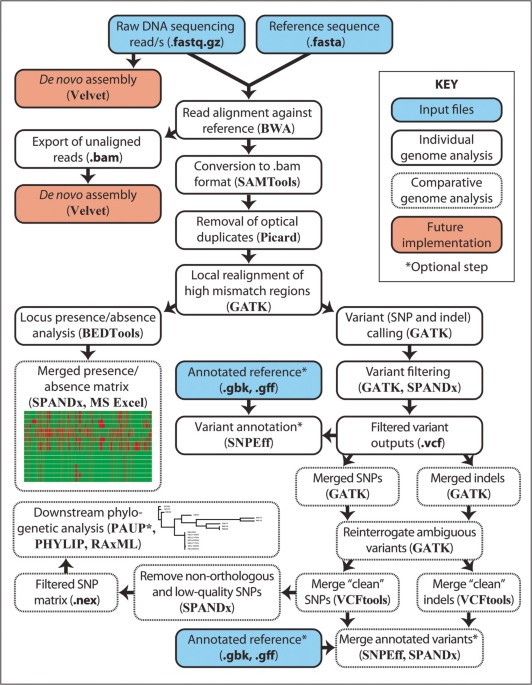
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes
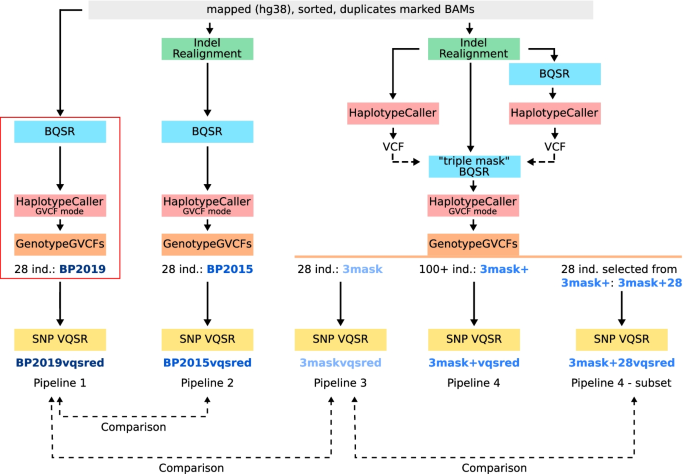
Comparison of sequencing data processing pipelines and application to underrepresented African human populations, BMC Bioinformatics
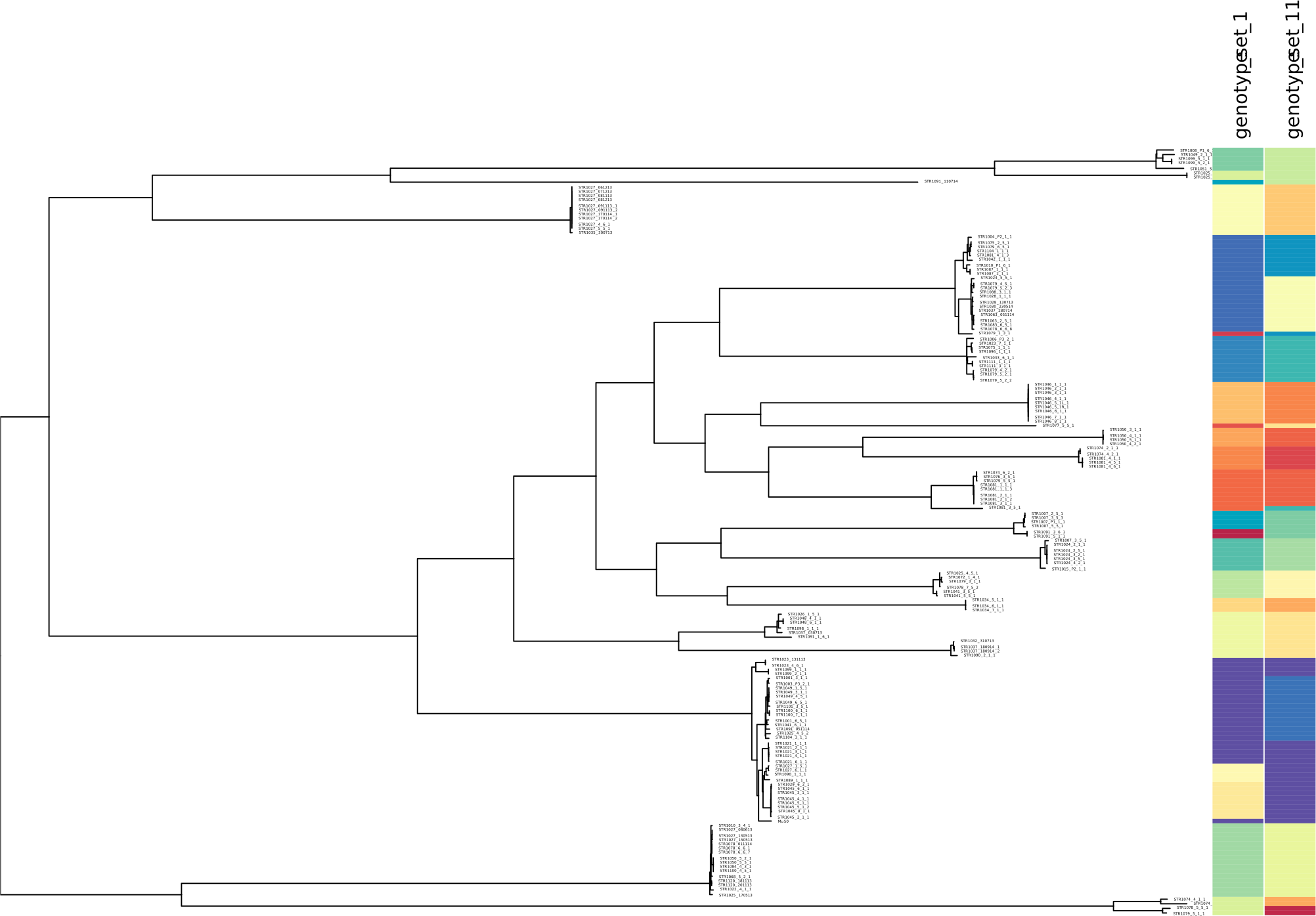
minSNPs: an R package for the derivation of resolution-optimised SNP sets from microbial genomic data [PeerJ]

Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality
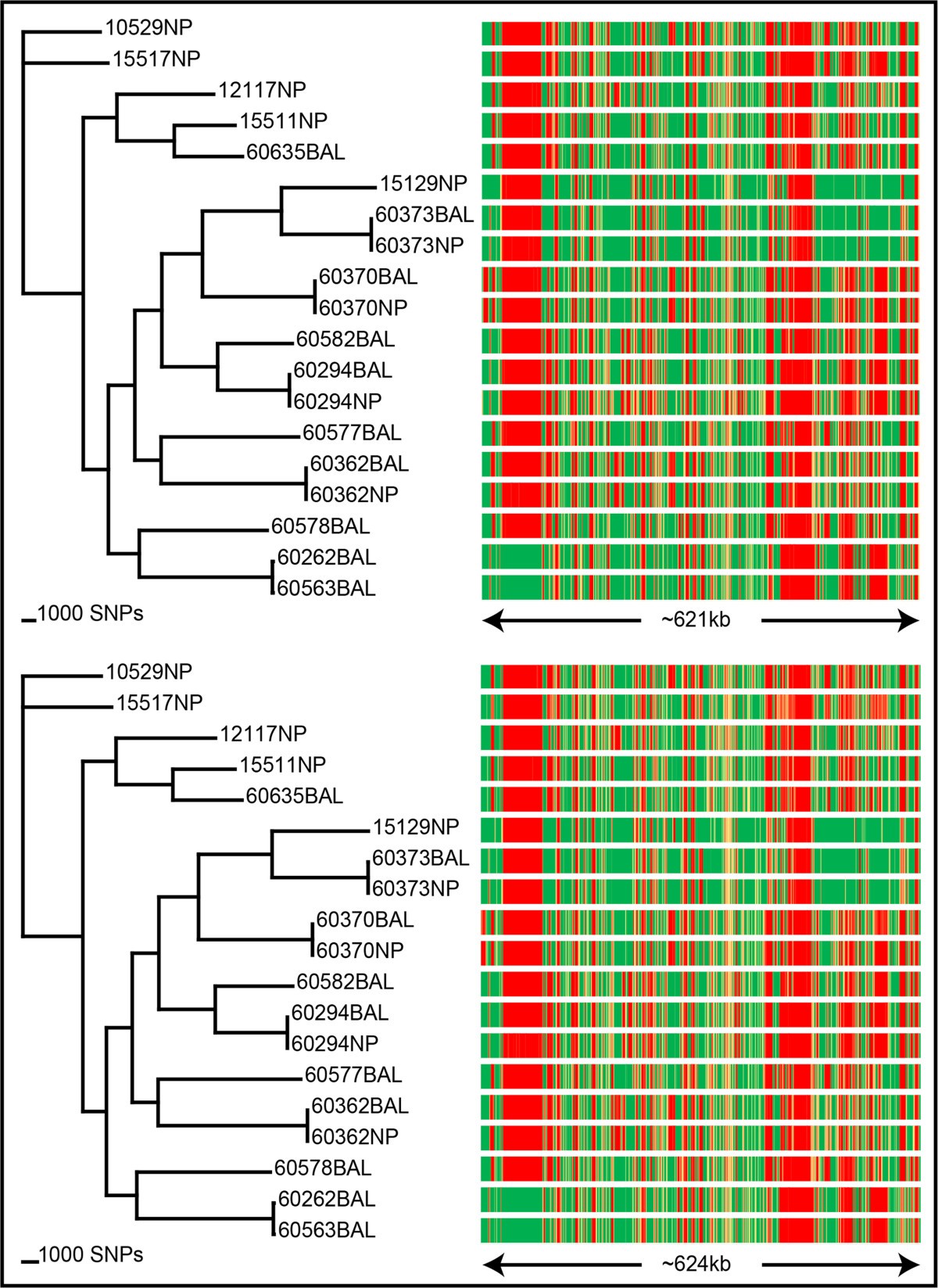
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes

Dynamics of genetic variation in Transcription Factors and its implications for the evolution of regulatory networks in Bacteria






