ICGC-TCGA DREAM Somatic Mutation Calling Challenge
🚧On this page:OverviewAccess ICGC-TCGA DREAM Somatic Mutation Calling Challenge resources on the CGCCopy the ICGC-TCGA DREAM Somatic Mutation Calling public projectLearn moreWebinar: Visual interfaceWebinar: Python and APIResources OverviewThe Seven Bridges CGC is proud to launch the ICGC-TCGA DREA

A challenge to the community to improve RNA sequencing data - Ontario Institute for Cancer Research

PDF) Combining accurate tumor genome simulation with crowdsourcing

NeoMutate: an ensemble machine learning framework for the prediction of somatic mutations in cancer, BMC Medical Genomics

precisionFDA/NCI-CPTAC Challenge Top Performers to Present at RECOMB2019 DREAM Satellite Conference

The ICGC-TCGA DREAM Somatic Mutation Calling Challenge Summary November 10, 2014 Dr. Paul C. Boutros Principal Investigator, Informatics & Biocomputing. - ppt download

(PDF) An ensemble approach to accurately detect somatic mutations

The Sentieon Genomic Tools - Improved Best Practices Pipelines for Analysis of Germline and Tumor-Normal Samples
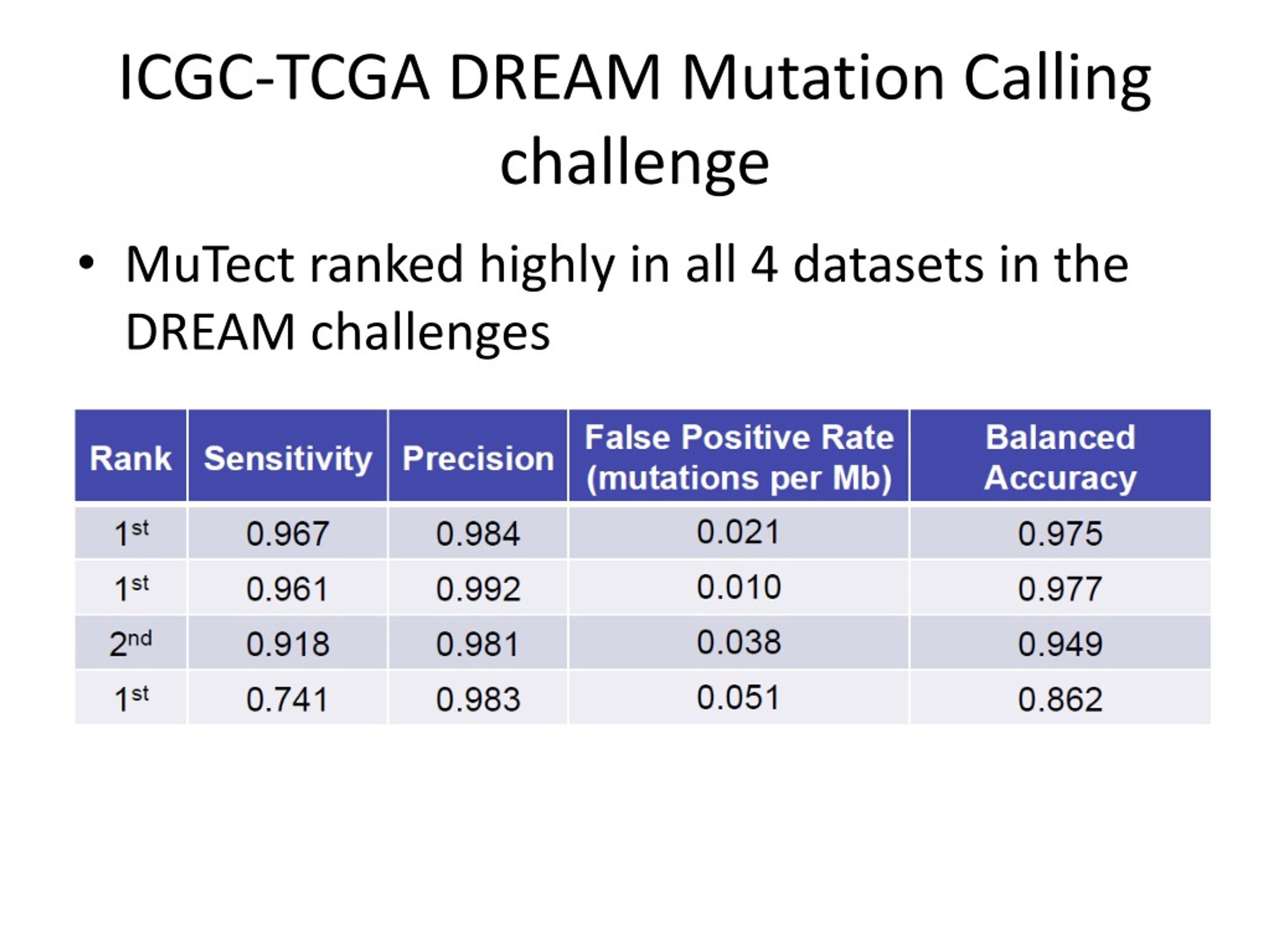
PPT - Data Analysis for Exome Sequencing Data PowerPoint Presentation, free download - ID:9068999

DREAM Challenge Stage 3 results trained from modified Stage 2 data
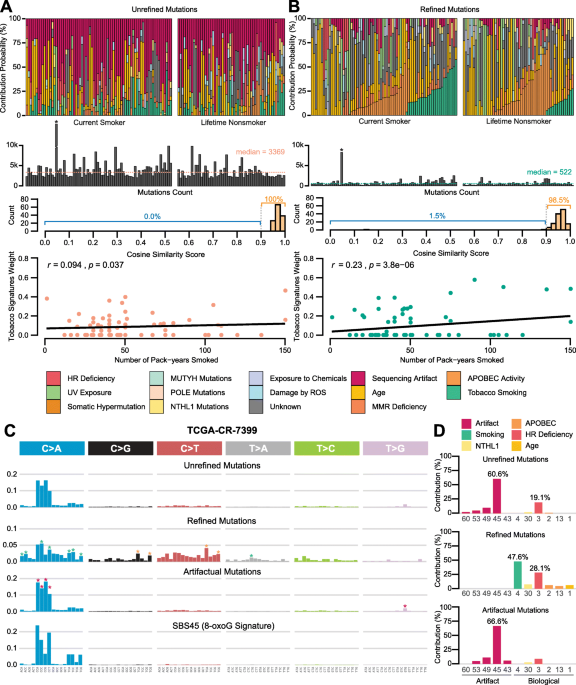
FIREVAT: finding reliable variants without artifacts in human

Optimizing novoBreak on the Cancer Genomics Cloud - SEVEN BRIDGES
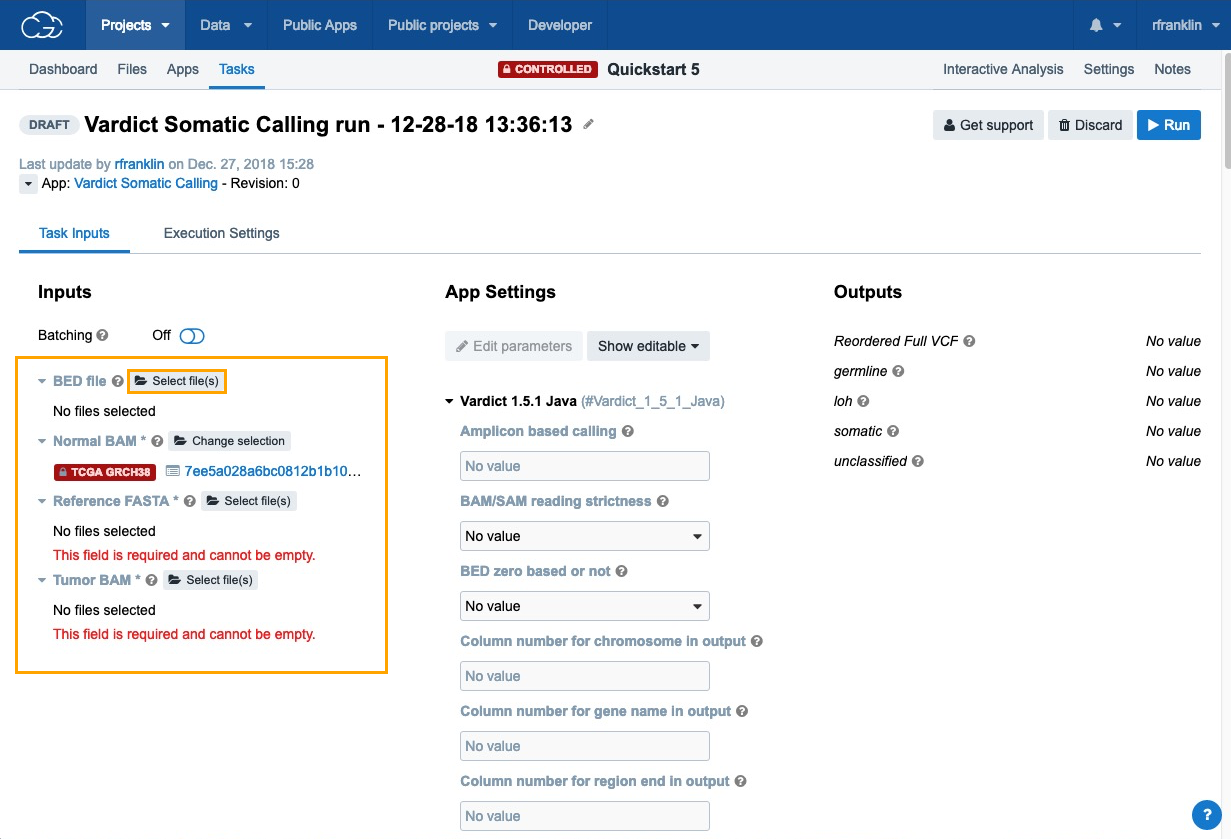
Quickstart - TCGA data (controlled access required)

1 Cancer Sequencing Quality & The ICGC-TCGA DREAM Somatic Mutation











