SPANDx - Comparative genomics for next-generation haploid sequence data - dsarov/SPANDx

Genomic diversity and antimicrobial resistance of Prevotella spp. isolated from chronic lung disease airways

Comparative genomics of Nocardia seriolae reveals recent importation and subsequent widespread dissemination in mariculture farms in South Central Coast, Vietnam
SPANDx download

Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality

Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of 'Haemophilus intermedius
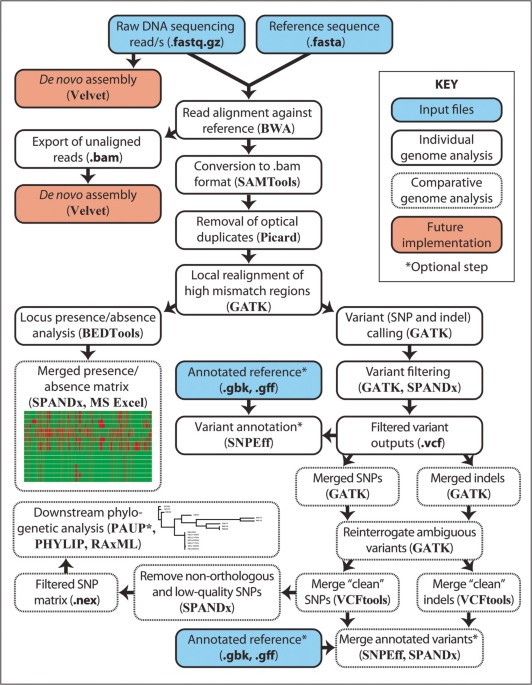
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes
GitHub - jeffersonfparil/compare_genomes: A comparative genomics workflow using Nextflow, conda, Julia and R

Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality

Degust volcano plots showing differentially expressed (DE) genes
GitHub - dsarov/SPANDx: SPANDx - Comparative genomics for next-generation haploid sequence data
Phylogeographic, genomic, and meropenem susceptibility analysis of Burkholderia ubonensis

Can non-typeable Haemophilus influenzae carriage surveillance data infer antimicrobial resistance associated with otitis media?
comparative-genomics · GitHub Topics · GitHub

Genomic Epidemiology Links Burkholderia pseudomallei from Individual Human Cases to B. pseudomallei from Targeted Environmental Sampling in Northern Australia
comparative-genomics · GitHub Topics · GitHub







